Chemoorganotrophy is a term used to denote the oxidation of organic chemicals to yield energy. In other words, an organic chemical serves as the initial electron donor. The process can be performed in the presence or absence of oxygen, depending upon what is available to a cell and whether or not they have the enzymes to deal with toxic oxygen by-products.
To start, let us focus on the catabolism of organic compounds when it occurs in the presence of oxygen. In other words, oxygen is being used as the final electron acceptor. When the process utilizes glycolysis and the tricarboxylic acid (TCA) cycle to completely oxidize an organic compound down to CO2, it is known as aerobic respiration. This generates the most ATP for a cell, given the large amount of distance between the initial electron donor (glucose) and the final electron acceptor (oxygen), as well as the large number of electrons that glucose has to donate.
In chemoorganotrophy, energy is derived from the oxidation of an organic compound. There are many different organic compounds available to a cell, such as proteins, polysaccharides, and lipids. But cellular pathways are arranged in such a way to increase metabolic efficiency. Thus, the cell funnels reactions into a few common pathways. By convention, glucose is used as the starting molecule to describe each process.
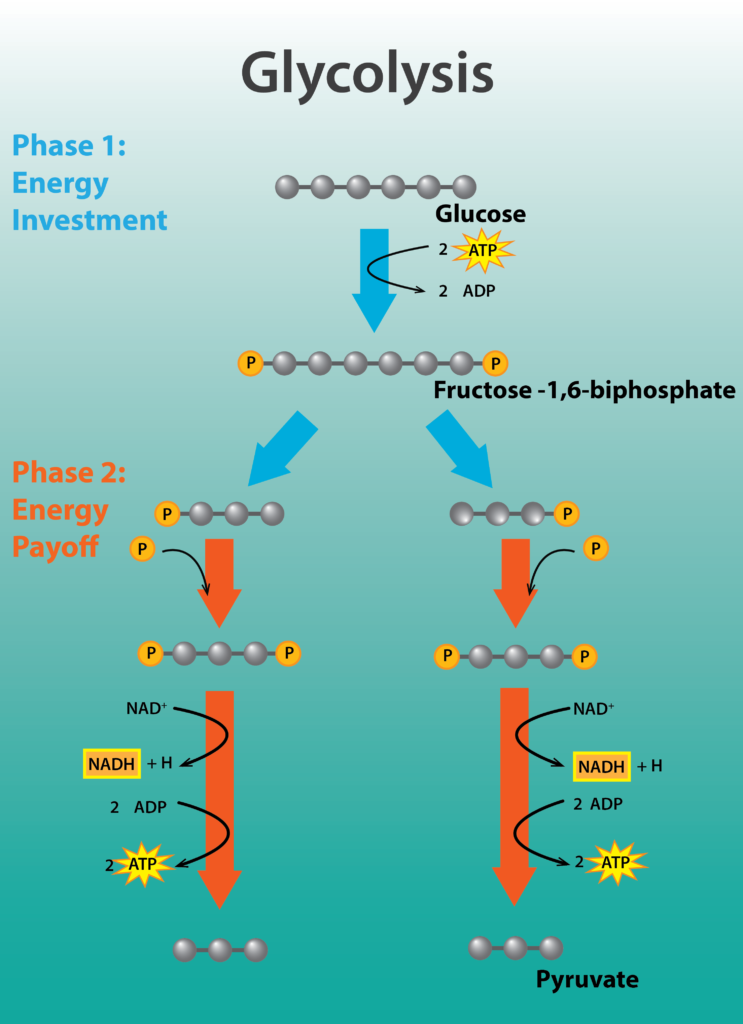
Glycolysis is a nearly universal pathway for the catabolism of glucose to pyruvate. The pathway is divided into two parts: part I, which focuses on modifications to the 6-carbon sugar glucose, and part II, where the 6-carbon compound is split into two 3-carbon molecules, yielding a bifurcated pathway. Part I actually requires energy in the form of 2 molecules of ATP, in order to phosphorylate or activate the sugar. Part II is the energy conserving phase of the reaction, where 4 molecules of ATP are generated by substrate-level phosphorylation, where a high-energy molecule directly transfers a Pi to ADP.
The net yield of energy from glycolysis is 2 molecules of ATP for every molecule of glucose. In addition, 2 molecules of the carrier NAD+ are reduced, forming NADH. In aerobic respiration, these electrons will ultimately be transferred by NADH to an electron transport chain, allowing the cell to capture more energy. Lastly, 2 molecules of the 3-carbon compound pyruvate are produced, which can be further oxidized to capture more energy for the cell.

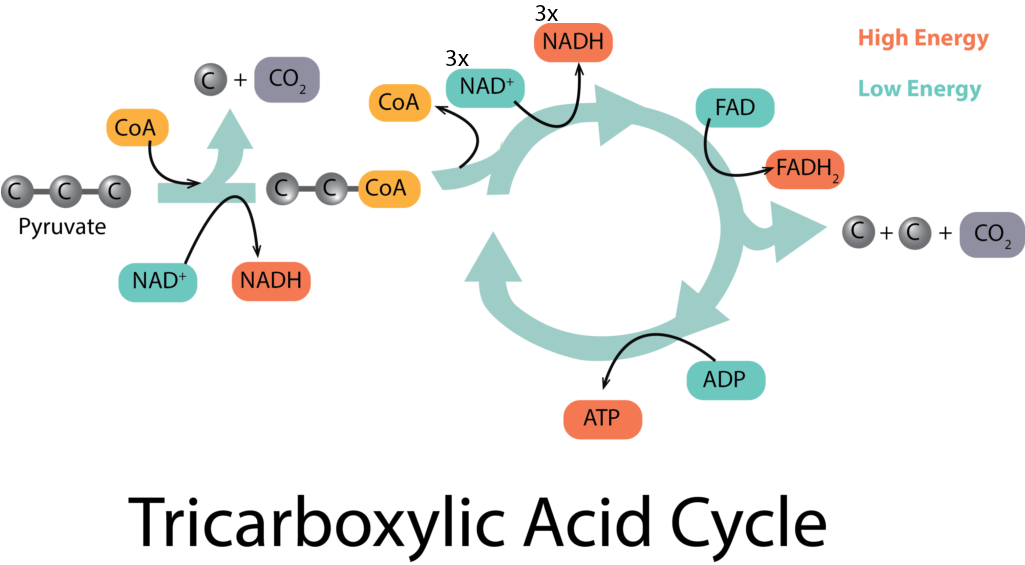
The tricarboxylic acid (TCA) cycle picks up at the end of glycolysis, in order to fully oxidize each molecule of pyruvate down to 3 molecules of CO2, as occurs in aerobic respiration. It begins with a type of connecting reaction before the molecules can enter the cycle proper. The connecting reaction reduces 1 molecule of NAD+ to NADH for every molecule of pyruvate, in the process of making citrate.
The citrate enters the actual cycle part of the process, undergoing a series of oxidations that yield many different products, many of them important precursor metabolites for other pathways. As electrons are released, carriers are reduced, yielding 3 molecules of NADH and 1 molecule of FADH2 for every molecule of pyruvate. In addition, 1 molecule of GTP (which can be thought of as an ATP-equivalent molecule) is generated by substrate-level phosphorylation.
Taking into account that there were two molecules of pyruvate generated from glycolysis, the net yield of the TCA cycle and its connecting reaction are: 2 molecules of GTP, 8 molecules of NADH, and 2 molecules of FADH2. But where does the ATP come from? So far we only have the net yield of 2 molecules from glycolysis and the 2 molecules of ATP-equivalents (i.e. GTP) from the TCA cycle. This is where the electron transport chain comes into play.
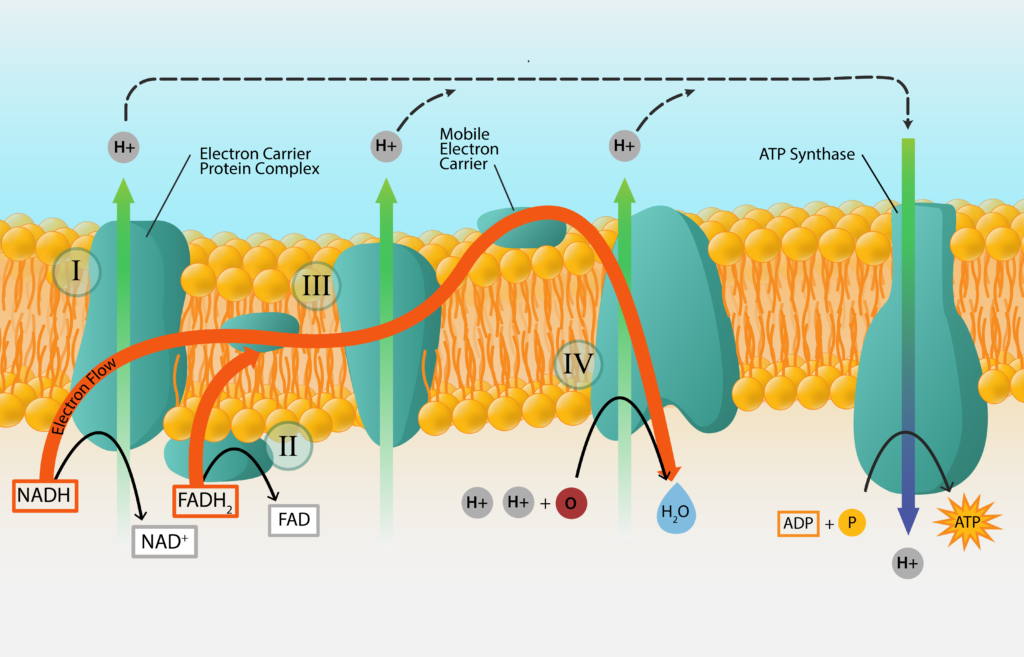
The synthesis of ATP from electron transport generated from oxidizing a chemical energy source is known oxidative phosphorylation. We have already established that electrons get passed from carrier to carrier, in order of their standard reduction potential. We have also established that some carriers accept electrons and protons, while others accept electrons only. What happens to the unaccepted protons? And how does this generate ATP for the cell? Welcome to the wonderful world of the proton motive force (PMF) and ATP synthase!
Protons that are not accepted by electron carriers migrate outward, to line the outer part of the membrane. For bacteria and archaea, this means lining the cell membrane and explains the importance for the negative charge of the cell.
As the positively charged protons accumulate, a concentration gradient of protons develops. This results in the cytoplasm of the cell being more alkaline and more negative, leading to both a chemical and electrical potential difference. This proton motive force (PMF) can be used to do work for the cell, such as in the rotation of the bacterial flagellum or the uptake of nutrients.
The PMF can also be used to synthesize ATP, with the help of an enzyme known as ATP synthase (or ATPase). This large enzyme has two components, one that spans the membrane and one that sticks into the cytoplasm and synthesizes the ATP. Protons are driven through the membrane-spanning component, generating torque that drives the rotation of the cytoplasmic portion. When the cytoplasmic component returns to its original configuration it binds Pi to ADP, generating a molecule of ATP.
After all that, what did the cell end up with, from using aerobic respiration? Using substrate-level phosphorylation the cell generated 2 net molecules of ATP during glycolysis, in addition to 2 molecules of ATP-equivalents from the TCA cycle. For reduced carriers, there were 2 molecules of NADH generated during glycolysis, in addition to 8 molecules from the TCA cycle or its connecting reaction. There were also 2 molecules of FADH2 from the TCA cycle. All of those electrons were passed on to the ETC (and eventually to oxygen), in order to develop a PMF, so that ATP synthase could generate ATP. How much ATP is generated?
Research indicates that the process is not completely efficient and there is some “leakage” that occurs. Current estimates are that 2.5 ATP are generated for every molecule of NADH, while 1.5 ATP are generated for every molecule of FADH2. Using these values would allow the cell to synthesize 25 molecules of ATP from all the NAD+ that was reduced in the process, in addition to 3 molecules of ATP from the FAD+ that was reduced. This would bring the grand total of maximum ATP produced to 32 (counting the GTP in that figure).
Certainly oxygen is a wonderful final electron acceptor, particularly when paired with glucose as an initial electron donor. It is part of the lowest redox couple on an electron tower, with an extremely positive standard electron potential. But what does a microbe do, if oxygen is not available or it lacks the protections necessary from toxic oxygen by-products? Let us focus on the generation of energy in the absence of oxygen, using a different electron acceptor, when an organic chemical is still being used as the initial electron donor. Examples of anaerobic chemoorganotrophy include anaerobic respiration and fermentation.
Anaerobic respiration starts with glycolysis as well and the pyruvate can be shunted off to the TCA cycle, just like in aerobic respiration. In fact, oxidative phosphorylation is used to generate most of the ATP, which means the use of an ETC and ATP synthase. The key difference is that the final electron acceptor will not be oxygen.
There are a variety of possible final electron acceptors that can be used in anaerobic respiration, allowing microbes to live in a wide variety of locations. The best electron acceptor will be the one that is lowest down on the electron tower, in an oxidized form (i.e. on the left-hand side of the redox couple). Some common electron acceptors include nitrate (NO3-), ferric iron (Fe3+), sulfate (SO42-), carbonate (CO32-) or even certain organic compounds, like fumarate.
How much ATP is generated by anaerobic respiration? That will depend upon the final electron acceptor being used. It will not be as much as is generated during aerobic respiration, since we know that oxygen in the best possible electron acceptor. Selection of an electron acceptor other than oxygen pushes an organism up the electron tower, shortening the distance between the electron donor and the acceptor, reducing the amount of ATP produced.
No matter what they might teach you in a biochemical class, fermentation and anaerobic respiration are not the same thing, at least not to a microbiologist.
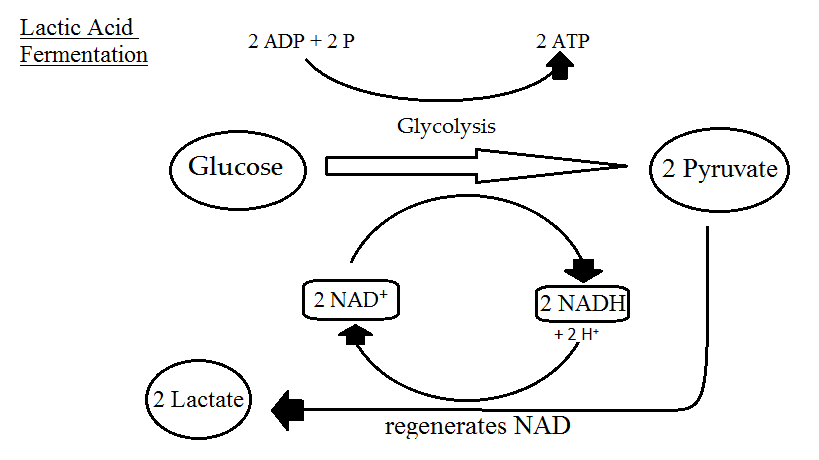
Fermentation is catabolism of glucose in the absence of oxygen as well and it does have some similarities to anaerobic respiration. Most obviously, it does not use oxygen as the final electron acceptor. It actually uses pyruvate, an organic compound. Fermentation starts with glycolysis, a process which we have already covered, that also starts off both aerobic respiration and anaerobic respiration. What does it yield? Two net molecules of ATP by substrate-level phosphorylation and 2 molecules of NADH. Organisms doing either aerobic or anaerobic respiration would then utilize oxidative phosphorylation in order to increase their ATP yield. Fermenters, however, lack an ETC or repress synthesis of their ETC when oxygen is not available, so they do not use the TCA cycle at all.
Without the use of an ETC (or a PMF or ATP synthase), no further ATP is generated beyond what was synthesized during glycolysis. But organisms using fermentation cannot just stop with glycolysis, since eventually all their molecules of NAD+ would become reduced. In order to re-oxidize this electron carrier they use pyruvate as a final electron acceptor, yielding a variety of fermentation products such as ethanol, CO2, and various acids.
Fermentation products, although considered waste products for the cell, are vitally important for humans. We rely on the process of fermentation to produce a variety of fermented foods (beer, wine, bread, cheese, tofu), in addition to using the products for a variety of industrial processes.
chemoorganotrophy, aerobic respiration, glycolysis, substrate-level phosphorylation, tricarboxylic acid (TCA) cycle, GTP, oxidative phosphorylation, proton motive force (PMF), ATP synthase/ATPase, anaerobic respiration, fermentation.
General Microbiology Copyright © 2019 by Linda Bruslind is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, except where otherwise noted.